Our Impact in Changing the World
At Complete Omics, we harness the power of proteomics and multi-omics assays to bridge groundbreaking scientific discovery with clinical excellence. Born at Johns Hopkins Medicine and growing globally, our journey from the first human proteome map to over 1 million sample analyzed underscores our commitment to innovation. With a legacy of over 600 publications and deep partnerships with leading research and clinical institutions, we’re redefining molecular diagnostics and personalized treatment. Join us in creating a healthier tomorrow,
TODAY
2023–Closing latest financing round with QiMing Venture Partners
Complete Omics proudly announces its latest financing round’s success, significantly backed by Qiming Venture Partners, China’s top healthcare VC. This achievement is particularly remarkable given the current economic climate, where securing investment has become increasingly challenging, especially in the healthcare and pharmaceutical sectors. Our team’s dedication and resilience have been key in reaching this pivotal moment for our company. Our collaboration with Qiming, known for investing in industry leaders like Xiaomi (1810.HK), Meituan (3690.HK), and Bilibili (BILI), signals strong confidence in our mission and future. This investment propels us into new realms of innovation in clinical proteomics and molecular diagnostics, underscoring our commitment to transforming healthcare. It positions us among influential firms, promising a future of growth and breakthroughs despite the economic slowdown. We’re excited for what’s ahead and invite our community to join us in this journey towards significant healthcare advancements. Together, we’re poised to redefine the landscape of personalized medicine. (Ref. 1, 2).A Proud Partner of QiMing VC2022–Direct Identification and Quantification of Neoantigens from Minute Amount of Clinical Biopsy Sample — published on Cancers
In recent years, neoantigens are becoming popular cancer therapeutic targets under intensive studies by almost all major oncology pharmaceuticals. However, who are the target patients? Does the patient with a “correct” mutation and a “correct” HLA allele indeed present the “correct” neoantigen? Is this individual’s neoantigen copy number high enough for immunotherapy? AI predictions based on NGS genomic information have been proven incapable of answering these questions. Immunopeptidome through mass spectrometry is dominated by disease-irrelevant peptide sequences. There is no existing way to identify and quantify neoantigens from a minute amount of clinical biopsy sample, such as 50 mg tissue or less. Valid-NEO is developed to fit this demanding clinical need through combining our proprietary multi-omics platforms including NGS-based ultra-rare mutation calling technique, DEEPER-SeqS, and our unique clinical proteomics platform, Complete360®, with additional hardware innovations (Ref. 1).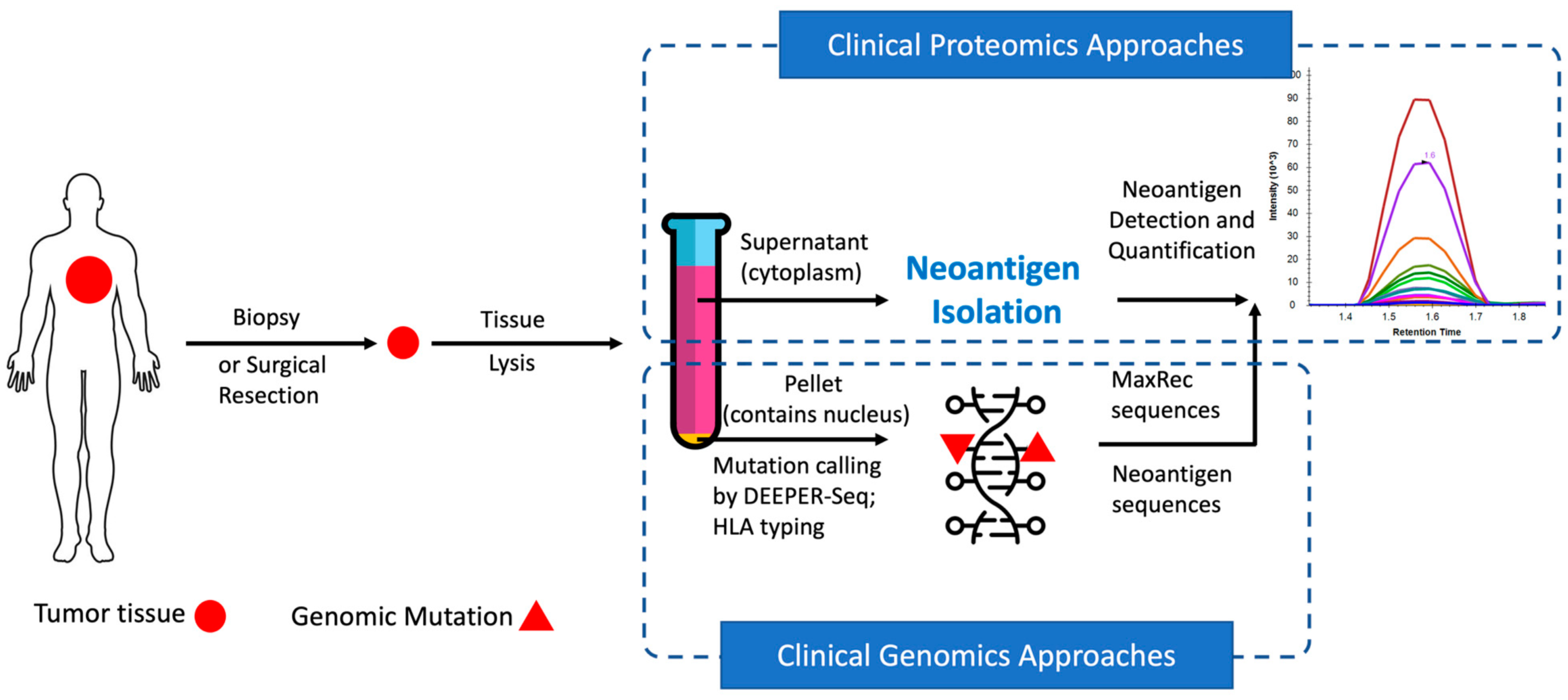 Valid-NEO: Multi-omics Pipeline for Neoantigen Assays
Valid-NEO: Multi-omics Pipeline for Neoantigen Assays2022–Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) — published on Science Advances
Human endogenous retroviruses (HERVs) represent 8% of the human genome. Working with, ErVaccine Technologies, our scientists identified neoantigens encoded by HERVs across a broad spectrum of cancers, and this finding may help enable next-generation therapeutic vaccines and cellular immunotherapies targeting these so-called “unconventional” tumor antigens (Ref 1). These antigens are shared by different tumor types. They would prove useful as personalized cancer therapeutic targets for a large number of patients. Along the side when we are keep on improving and developing our own disease detection and companion diagnostic pipelines, we are excited to be working with a large number of collaborators to implement our multi-omics platforms in their different clinical and basic research projects.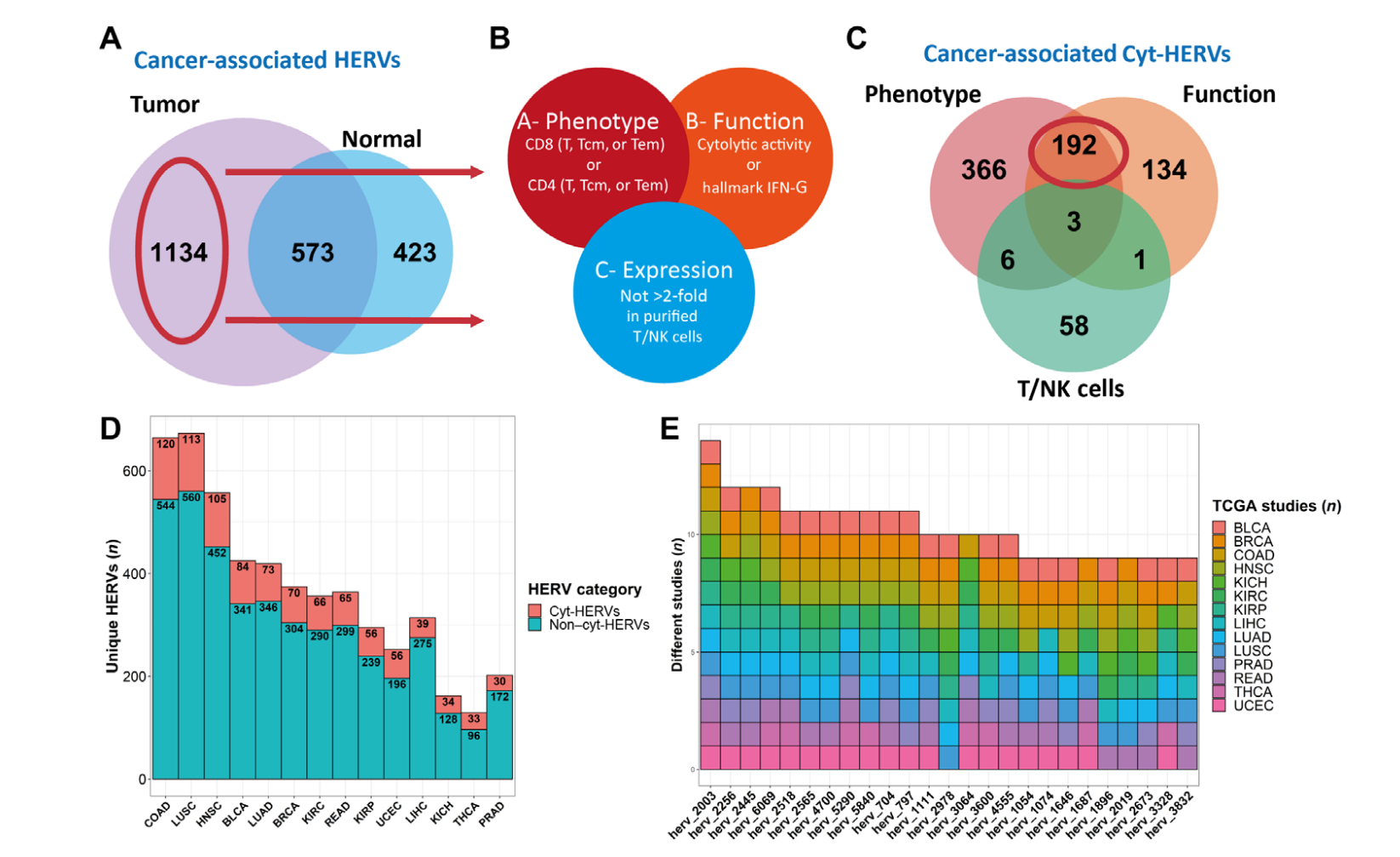 Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) — A new class of cancer therapeutic targets
Identify and Quantify Neoantigens derived from Human endogenous retroviruses (HERVs) — A new class of cancer therapeutic targets2021–Therapeutic Neoantigens Encoded by Oncogene K-Ras — published on Science Immunology
K-Ras is one of the most highly mutated oncogenes in cancers. The neoantigens encoded by K-Ras can be presented by many different types of cancer cells. Here we utilized our multi-omics neoantigen validation pipeline to detect and quantify K-Ras neoantigens from a variety of cancer samples (Ref 1.). We found that the K-Ras as well as several other oncogenes can be presented on cancer cell surface, but at extremely low copy numbers. We adopted a variety of internal control system to measure their abundance down to <1 copy per cell level. To come up with an actionable strategy, the therapeutic team at JHU developed bispecific antibodies that can target the neoantigens we identified, and deliver dramatic therapeutic effects to mice. These findings are significant in the way that it opens a gate to developing pan-cancer immunotherapeutic agents that can treat a large number of patients sharing cancer hotspot mutations. Identifying such neoantigens is the first step in this campaign and is being accomplished by Complete Omics Inc.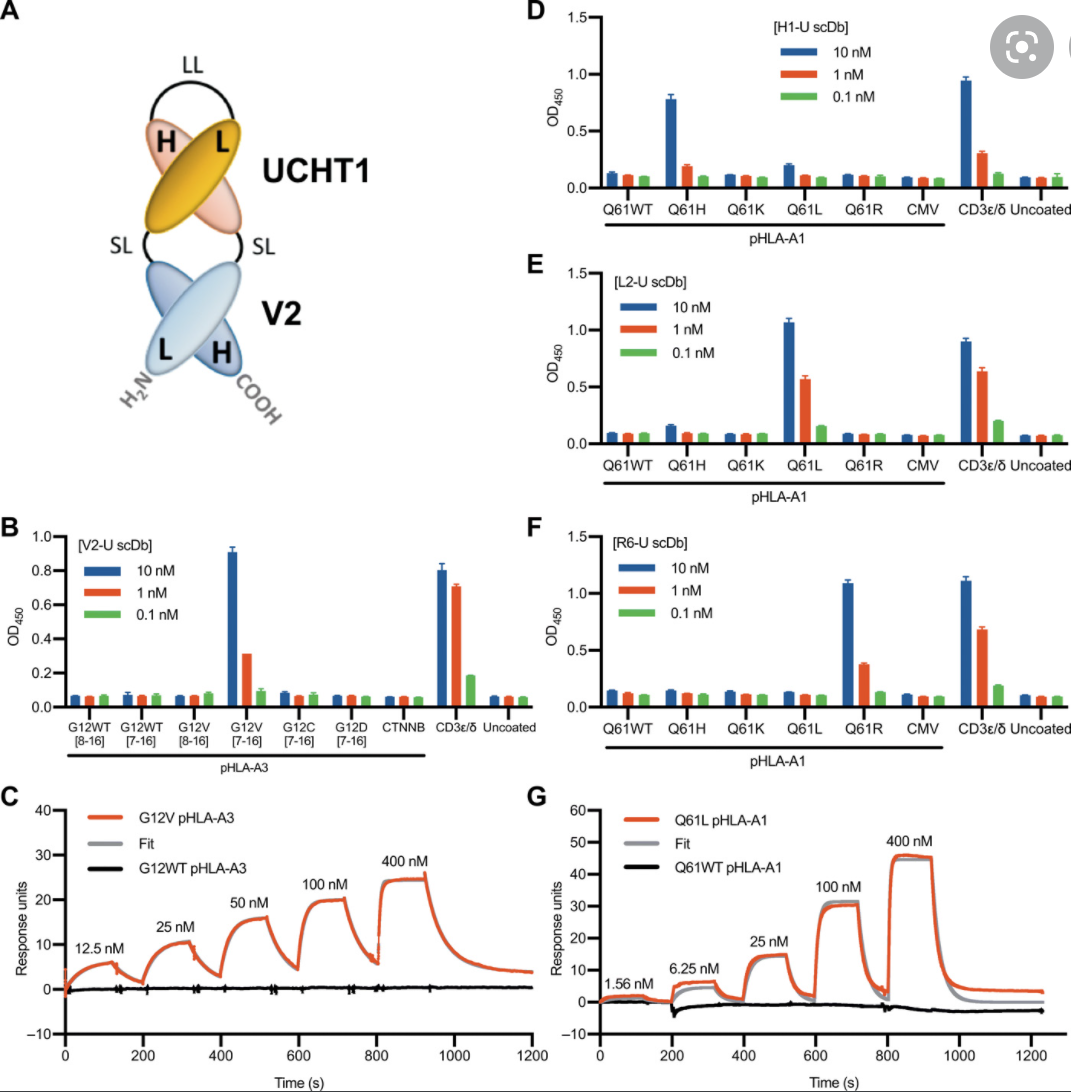 K-Ras Neoantigen Identified for Personalized Cancer Therapeutics
K-Ras Neoantigen Identified for Personalized Cancer Therapeutics2021–Direct Quantification of Neoantigens Like Never Before — published on Science
Genetic changes in human genome are the driving force for all cancers. Different patients have different sets of mutation profiles even for the patients who all have the same disease. For decades, doctors, cancer researchers, and pharmaceutical companies have been working tirelessly trying to find a way to treat each person’s unique disease in a highly personalized way that will reach the maximum treatment efficacy with the lowest side effects. Complete Omics, working with leaders in cancer therapeutics, has developed pipelines based on our multi-omics techniques through which we clearly observe and quantify personalized therapeutic targets encoded by the most frequently mutated tumor suppressor gene TP53. We validated and quantified the TP53 neoantigens on the surfaces of cancers and provided information to healthcare providers to support their decision on if or not to adopt a highly personalized cancer treatment targeting these neoantigens and when to use it (Ref 1). These findings provided the 1st-hand evidence for cancer therapeutics without the uncertainty that comes with predictions.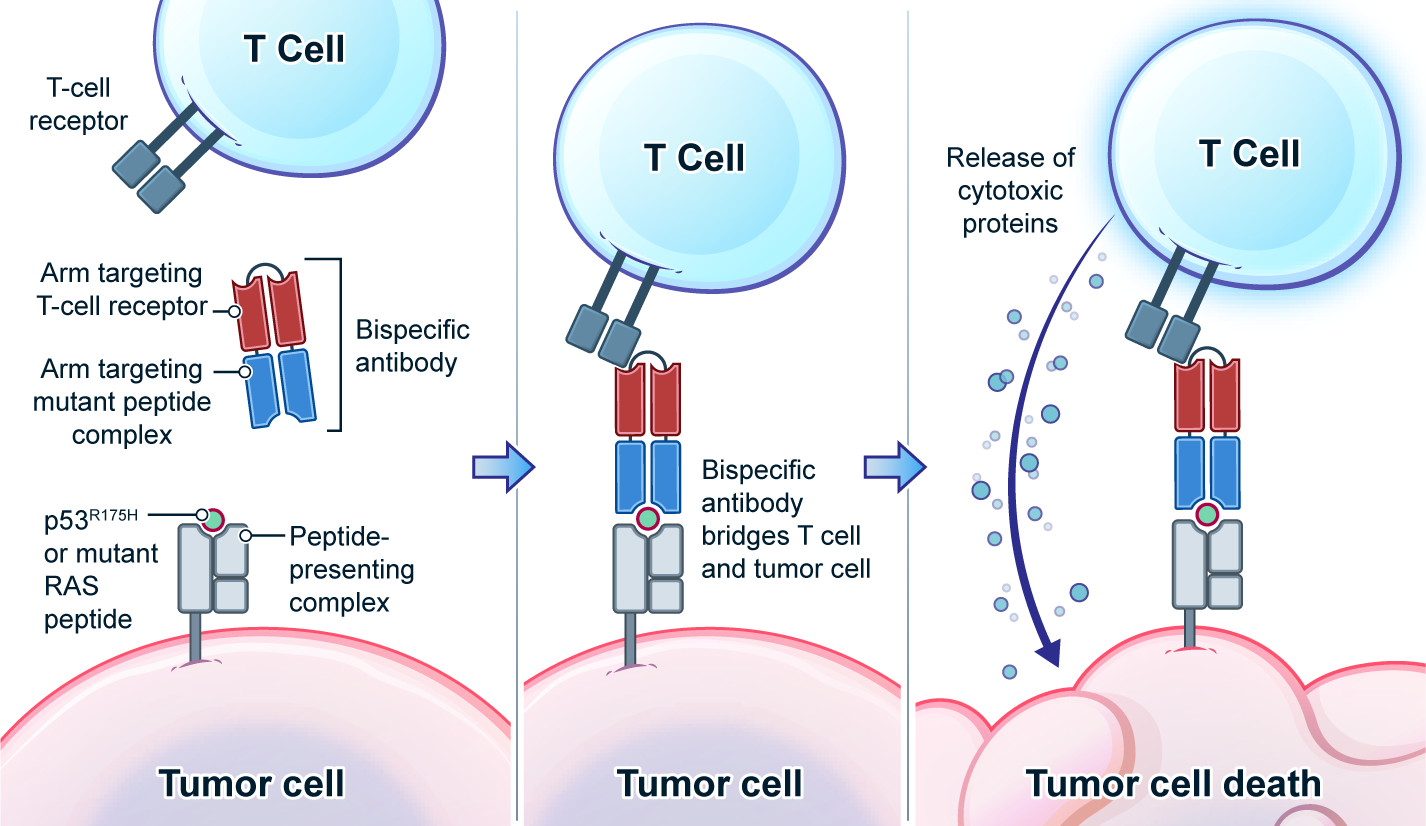 Prediction-FREE Neoantigen Validation Enables TP53-targeted Personalized Cancer Therapeutics
Prediction-FREE Neoantigen Validation Enables TP53-targeted Personalized Cancer Therapeutics2019–First Pipeline for Direct Detection and Quantification of Neoantigens — published on Cancer Immunology Research
Cancer-linked genetic mutations can code for mutant proteins, which can then be processed by proteasomes into peptides that are presented by human leukocyte antigen (HLA) molecules, triggering the body’s immune response. The idea that such mutant peptides can trigger an immune response is fundamental to immunotherapies like checkpoint inhibitors as well as cancer vaccines that present the body with these peptides to generate an immune reaction. However, while the rise of next-generation sequencing has allowed researchers to identify a large number of cancer-linked mutations, actually detecting these mutation-associated neo-antigens, or MANAs, at the peptide level remains difficult. The fact that a mutant is present at the genetic level does not mean it will be produced at the protein level, and, even if it is, that is no guarantee that it will be processed and presented by HLA molecules. This has proved a challenge for, for instance, personalized cancer vaccine development. Our technology (Ref. 1) provide the 1st pipeline for detecting personalized cancer therapeutic targets. Our method has been reported by public media and has a significant impact in cancer research (Ref. 2).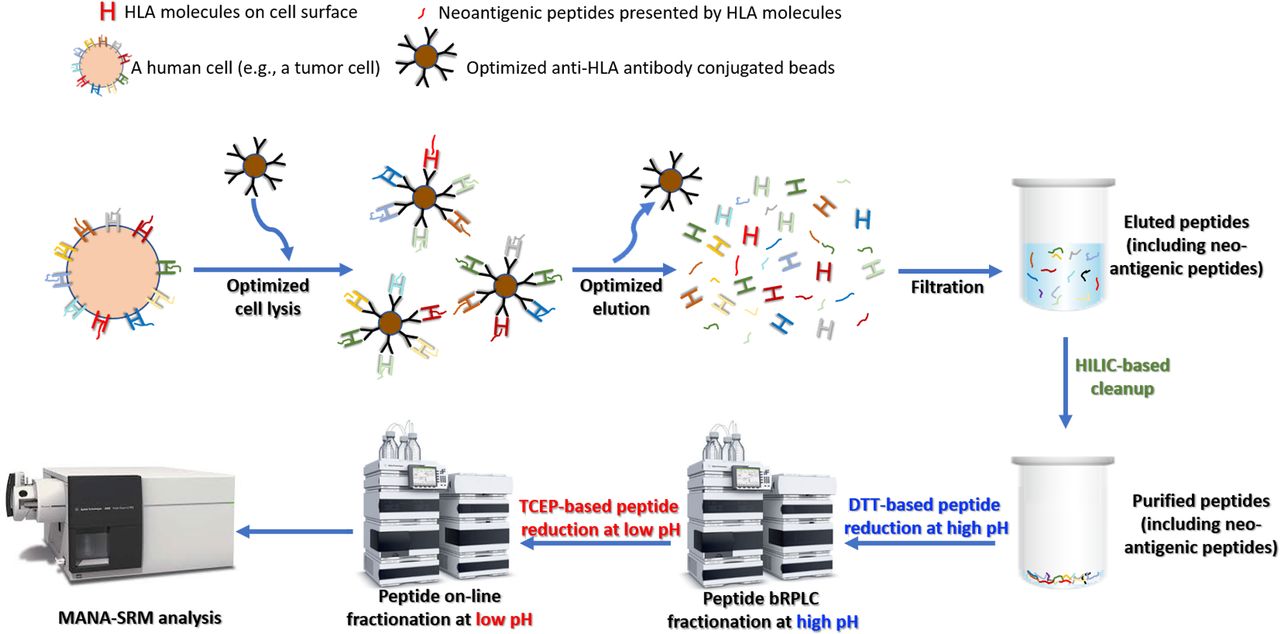 Direct Detection and Quantification of Neoantigens
Direct Detection and Quantification of Neoantigens2017–First Pipeline for Proteomics-based Clinical Diagnostics — published on PNAS
We developed the 1st pipeline for discovering and validating proteomic biomarkers from patient plasma samples (Ref 1). With this pipeline, we identified an ovarian cancer biomarker, peptidyl-prolyl cis–trans isomerase A (PPIA), with zero false positive diagnostic value. Our finding was featured on Genomeweb (Ref 2). We quantified 318 peptides in 94 separate samples, 48 from healthy controls, 14 from colorectal cancer cases, 14 from ovarian cancer cases, and 18 from pancreatic cancer cases. We then analyzed this data to see if any of the peptides or combinations of the peptides allowed us to accurately identify their sample of origin, and in this way identified the two PPIA peptides as potential markers for ovarian cancer. We then measured these two peptides in a separate cohort of 73 cases consisting of 35 ovarian cancer cases and 14 healthy controls and 24 pancreatic cancer cases. One of the two peptides was detected in 20 of the 35 ovarian cancer plasma samples, while neither was detected in any of the healthy controls.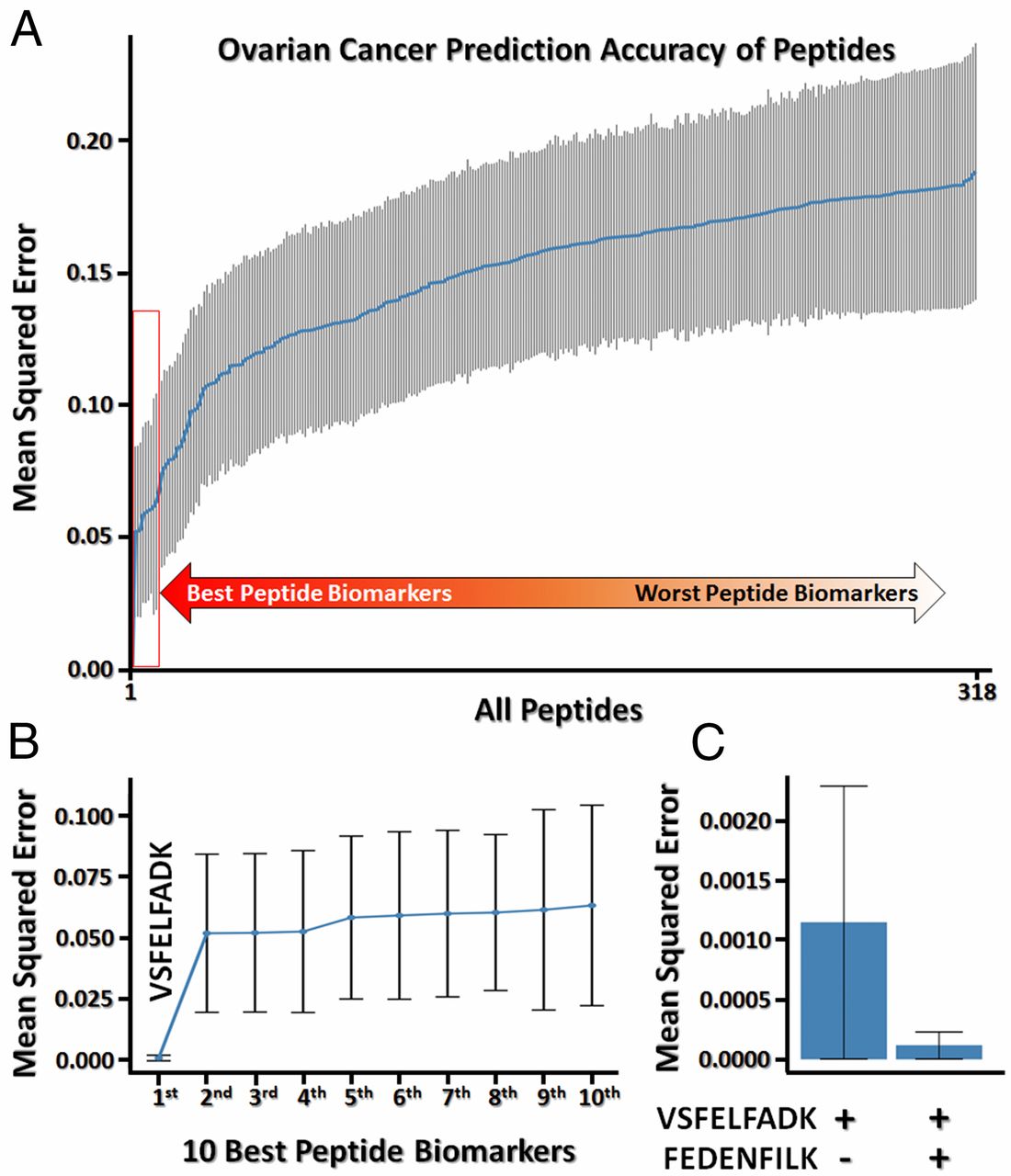 SAFE-SRM and Ovarian Cancer Diagnostics
SAFE-SRM and Ovarian Cancer Diagnostics2016–Targeted sequencing of both DNA strands barcoded and captured individually by RNA probes to identify genome-wide ultra-rare mutations — published on Scientific Reports
Targeted sequencing has been an essential application in Next Generation Sequencing (NGS). However, all target capture commercial technologies on the market have relatively low capturing efficiencies. That being said, after utilizing those capture kits, instead of sequencing a sufficient number of unique molecules, you are actually sequecing a library of nucleic acid amplicons that are amplified from very few DNA molecules captured by those old commercial technologies from initial NGS library, and usually the fraction of molecules that can be captured and subsequently sequenced is less than 9% of the molecules from the initial NGS library. Due to the unsatisfying performance of current commercial capture kits and platforms, Complete Omics has been trying to develop its own target capture technologies, and finally we have something to show you. We believe that this new technology could potentially revolutionize NGS targeted sequencing field. We are proud to anounce DEEPER-Capture, the next generation target capture technology for NGS sequencing. In DEEPER-Capture both DNA strands from the same DNA duplex molecule are captured by a pair of complementary RNA probes, and sequenced, simutanously. Using DEEPER-Capture, we repeatedly achieved an over 3-fold higher capturing efficiency than the best performance ever observed from competing platforms reported by numerous labs.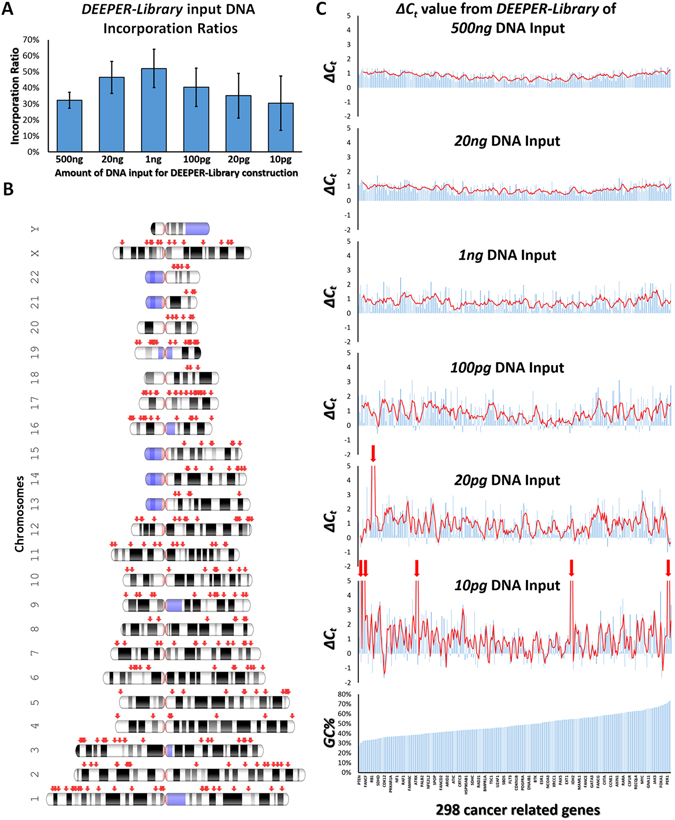 DEEPER-Seq: Capture both DNA strands with dual RNA probes
DEEPER-Seq: Capture both DNA strands with dual RNA probes2010–Mutant Proteins As Cancer Specific Biomarkers — published on PNAS
Cancer biomarkers are currently the subject of intense research because of their potential utility for diagnosis, prognosis, and targeted therapy. In theory, the gene products resulting from somatic mutations are the ultimate protein biomarkers, being not simply associated with tumors but actually responsible for tumorigenesis. In 2010, we developed the 1st pipeline for detecting and quantifying mutant protein as cancer specific biomarkers from human specimens, and it received a broad range of attentions from different disciplines of biomedical sciences [Ref. 1]. We demonstrated that altered protein products resulting from somatic mutations can be identified directly and quantified. As a prototypical example of this approach, we demonstrated that it is possible to quantify the number and fraction of mutant Ras protein present in cancer cell lines. There were an average of 1.3 million molecules of Ras protein per cell, and the ratio of mutant to normal Ras proteins ranged from 0.49 to 5.6. Similarly, we found that mutant Ras proteins could be detected and quantified in clinical specimens such as colorectal and pancreatic tumor tissues as well as in premalignant pancreatic cyst fluids. In addition to answering basic questions about the relative levels of genetically abnormal proteins in tumors, this approach lays the foundation for mutant protein-based cancer diagnostics.- Publication: Mutant proteins as cancer-specific biomarkers.
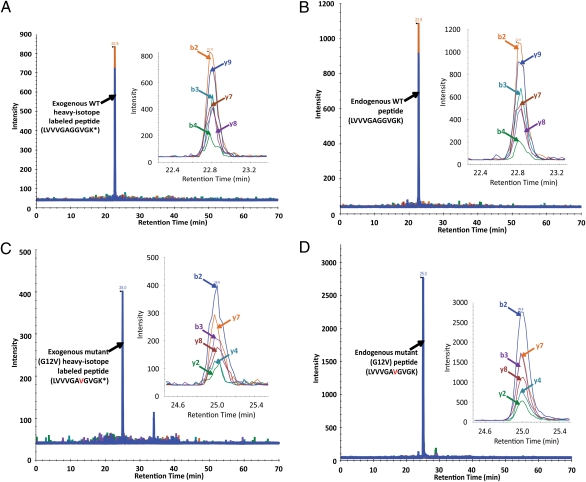 Mutant Proteins As Cancer Specific Biomarkers
Mutant Proteins As Cancer Specific Biomarkers